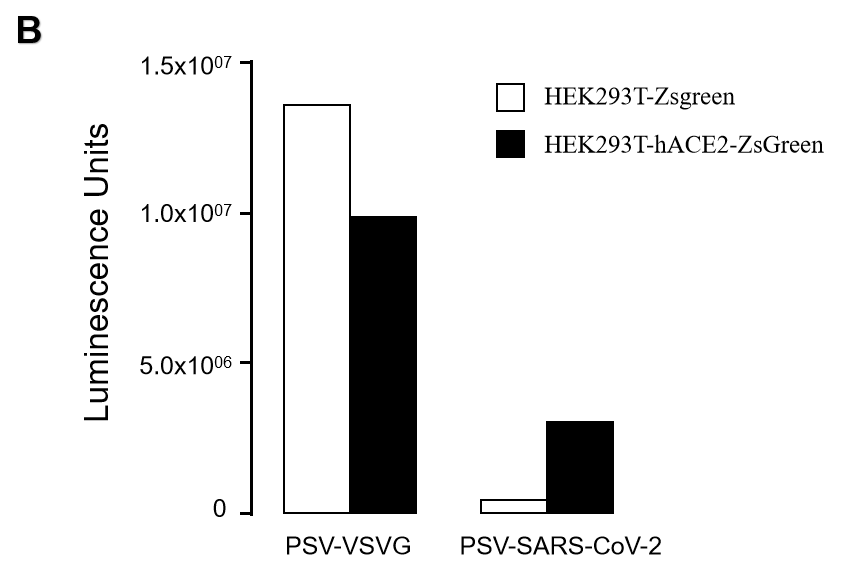Products & solutions & information collection of COVID-19 vaccines, new variants of SARS-CoV-2 and efficacy evaluation solutions
Diagnostic antibodies and antigens for Companion Animal disease testing
● Rabbit
Diagnostic antibodies and antigens for Swine disease testing
Diagnostic antibodies and antigens for Avian disease testing
Diagnostic antibodies and antigens for Multiple animal disease testing
Diagnostic antibodies and antigens for Ruminant disease testing
● Deer
Diagnostic antibodies and antigens for infectious and non-infectious Equine/Horse disease testing
SOCAIL MEDIA

Table of Contents
BACKGROUND
Challenge of COVID-19 vaccine discovery & development: to meet accumulated mutating of SARS-CoV-2 and a long-term viral genome transcrption
The efficacy and safety of COVID-19 vaccine candidates need carefully investigated, from pre-clinical, clinal stage to a long time after BLA. There are 2 important items here:
Firstly, the coronavirus is kept accumulated mutating. The most important mutation occurs in SARS-CoV-2 (2019nCoV) Spike protein (SARS-CoV-2 S protein). The SARS-CoV-2 (2019nCoV) Spike mediates binding and entry into host cells and is a major target of neutralizing antibodies. Most of the COVID-19 vaccines focus on spike protein1,4-6.
Different SARS-CoV-2 lineages with diverse Spike protein mutant variants may yield a heavy impact on the course of the pandemic2-3. The United Kingdom (UK) has faced a rapid increase in COVID-19 cases caused by a novel SARS-COV-2 (2019nCOV) lineage, the B.1.1.7 lineage, which carries a larger than a usual number of coronavirus genetic changes7-8, particularly in the SARS-CoV-2 spike protein(Table 1)7.
| Gene | Nucleotide | Amino acid | |
| ORF1ab | C3267T | T1001I | |
| C5388A | A1708D | ||
| T6954C | I2230T | ||
| 11288-11296 deletion | SGF 3675-3677 deletion | ||
| spike | 21765-21770 deletion | HV 69-70 deletion (Click to more details about HV 69-70 deletion related products) | |
| 21991-21993 deletion | Y144 deletion | ||
| A23063T | N501Y (Click to more details about N501Y related products) | ||
| C23271A | A570D | ||
| C23604A | P681H | ||
| C23709T | T716I | ||
| T24506G | S982A | ||
| G24914C | D1118H | ||
| Orf8 | C27972T | Q27stop | |
| G28048T | R52I | ||
| A28111G | Y73C | ||
| N | 28280 GAT->CTA | D3L | |
| C28977T | S235F |
Secondly, SARS-CoV-2 was reported to integrate into the genome, which means a stable long-term expression of transcript products of SARS-CoV-2 9.
In conclusion, the Efficacy of COVID-19 vaccines and neutralized antibodies need long-term tracking, which mainly focuses on the novel spike protein mutation occurring in the new lineage variant of SARS-CoV2. The efficacy evaluation solution and tools of COVID-19 vaccines and neutralized antibodies need continually updated.
PRODUCTS & PROCOTOL COLLECTION
GeneMedi Efficacy Assay/Evaluation Solutions for COVID-19 Vaccines and Therapeutic antibodies against SARS-CoV2(2019nCoV)
To meet accumulated mutating of SARS-CoV-2, GeneMedi keeps continually developing novel solutions and tools for efficacy evaluation of COVID-19 vaccines (and neutralized antibodies) against novel mutant SARS-CoV-2 (2019nCoV) lineages. Our scientist’s team takes duty in fighting against the COVID-19 pandemic with our global industrial and academic partners.
GeneMedi information and products collection of SARS-COV-2 (2019nCOV) UK B.1.1.7 lineage and South Africa 501Y.V2 lineage and Brazilian P.1 lineage(B.1.1.28.1)
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) B.1.1.7 lineage
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) B.1.1.7 lineage, which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Cat No. | Description | Order
|
|---|---|---|
Spike of SARS-COV-2 B.1.1.7 lineage mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2) N501Y mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2)-HV 69-70 Deletion mutation(ΔH69/ΔV70) SARS-CoV-2(2019nCoV) Pseudotyped virus |
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) 501Y.V2 lineage
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) S501Y.V2 lineage, which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Cat No. | Description | Order
|
|---|---|---|
Spike D614G mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2) N501Y mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2)-E484K mutation of SARS-CoV-2(2019nCoV) Pseudotyped virus (501Y.V2 lineage) |
||
Spike (S1+S2) RBD triple mutation(K417N, E484K and N501Y) of SARS-CoV-2(2019nCoV) Pseudotyped virus (501Y.V2 lineage) |
||
Spike (S1+S2) whole mutation variant of SARS-CoV-2(2019nCoV) 501Y.V2 lineage Pseudotyped virus |
GeneMedi pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) P.1 lineage(B.1.1.28.1, Brazilian variant)
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) P.1 lineage(B.1.1.28.1, Brazilian variant), which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Cat No. | Description | Order
|
|---|---|---|
Spike D614G mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2) N501Y mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike(S1+S2)-E484K mutation of SARS-CoV-2(2019nCoV) Pseudotyped virus (501Y.V2 lineage) |
||
Spike (S1+S2) RBD triple mutation(K417N, E484K and N501Y) of SARS-CoV-2(2019nCoV) Pseudotyped virus (501Y.V2 lineage) |
||
Spike (S1+S2) whole mutation variant of SARS-CoV-2(2019nCoV) 501Y.V2 lineage Pseudotyped virus |
SARS-COV-2 (2019nCOV) B.1.1.7 lineage & 501Y.V2 lineage(B.1.351) & P.1 lineage (B.1.1.28.1, Brazilian variant) of Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
Cat No. | Products Name | Source (Expression Host) | Tag | Bioactivity validation | Order
|
|---|---|---|---|---|---|
Recombinant human soluble ACE2 protein (soluble hACE2,extracellular hACE2,C-His) |
Mamamlian (human cell) |
C-His |
Spike(S-RBD) protein binding |
||
Recombinant human soluble ACE2 protein (soluble hACE2,extracellular hACE2,C-FC) |
Mamamlian (human cell) |
C-Fc |
Spike(S-RBD) protein binding |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike S-trimer protein D614G mutant (S1+S2-D614G trimer,C-6His) with furin cleavage site mutation & T4 fibritin trimerization motif |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike Protein S1 D614G mutant (S1-D614G,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) (UK variant B.1.1.7 lineage)
Spike Protein RBD-N501Y mutant (SRBD-N501Y,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) (South African variant 501Y.V2 lineage,B.1.351)
Spike Protein RBD(K417N, E484K and N501Y mutant) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) P.1 lineage(B.1.1.28.1, Brazilian variant)
Spike RBD Protein(K417T, E484K and N501Y mutant) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) B.1.1.7 lineage
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) B.1.1.7 lineage.
Cat No. | Description | Vector | Tag | Coden Optimized | Order
|
|---|---|---|---|---|---|
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
|||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
|||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
|||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) South Africa 501Y.V2 lineage(B.1.351).
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) S501Y.V2 lineage.
Cat No. | Description | Vector | Tag | Coden Optimized | Order
|
|---|---|---|---|---|---|
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
|||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) - P.1 lineage(B.1.1.28.1, Brazilian variant)
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) P.1 lineage(B.1.1.28.1, Brazilian variant)
Cat No. | Description | Vector | Tag | Coden Optimized | Order
|
|---|---|---|---|---|---|
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
|||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
pcDNA3.1(+) |
No tag |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
||
Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 8 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 76 Warning: Undefined variable $field_name in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 77 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 89 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 98 Warning: Trying to access array offset on value of type bool in /www/wwwroot/genemedi_com0630/wp-content/plugins/wc-product-table-pro/templates/custom_field.php on line 105 |
Pre-made adenovirus |
C-3FLAG |
coden optimized
for mamamlian |
Multiple variants of Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
The Spike proteins variants that GeneMedi offer is including:
1) Multiple variants of recombinant Spike protein for Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
Cat No. | Products Name | Source (Expression Host) | Tag | Bioactivity validation | Order
|
|---|---|---|---|---|---|
Recombinant 2019-nCoV(SARS-CoV-2) Spike S-trimer protein(S1+S2 trimer,C-6His) with furin cleavage site mutation (R682G,R683S,R685S) & T4 fibritin trimerization motif |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike S-trimer protein D614G mutant (S1+S2-D614G trimer,C-6His) with furin cleavage site mutation & T4 fibritin trimerization motif |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike S-trimer protein S6P mutant (F817P,A892P,A899P,A942P,K986P,V987P) with furin cleavage site mutation (R682G,R683S,R685S) & T4 fibritin trimerization motif |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike S-trimer Protein (Omicron variant aka B.1.1.529 lineage) with furin cleavage site mutation & T4 fibritin trimerization motif |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike Protein S1 D614G mutant (S1-D614G,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) (UK variant B.1.1.7 lineage)
Spike Protein RBD-N501Y mutant (SRBD-N501Y,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) (South African variant 501Y.V2 lineage,B.1.351)
Spike Protein RBD(K417N, E484K and N501Y mutant) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike Protein RBD V341I mutant (SRBD-V341I,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike Protein RBD F342L mutant (SRBD-F342L ,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
||
Recombinant 2019-nCoV(SARS-CoV-2) Spike Protein RBD N354D mutant (SRBD N354D ,C-HisTag) |
Mamamlian (human cell) |
C-His |
ACE2 binding;
Immunogen in Sandwich Elisa, lateral-flow tests,and other immunoassays; |
2) Recombinant human ACE2 protein products
Cat No. | Antigen Name of 2019-nCoV(SARS-CoV-2) | Isotypes | Order |
|---|---|---|---|
Recombinant human soluble ACE2 protein (soluble hACE2,extracellular hACE2,C-His) |
Mamamlian (human cell) |
||
Recombinant human soluble ACE2 protein (soluble hACE2,extracellular hACE2,C-FC) |
Mamamlian (human cell) |
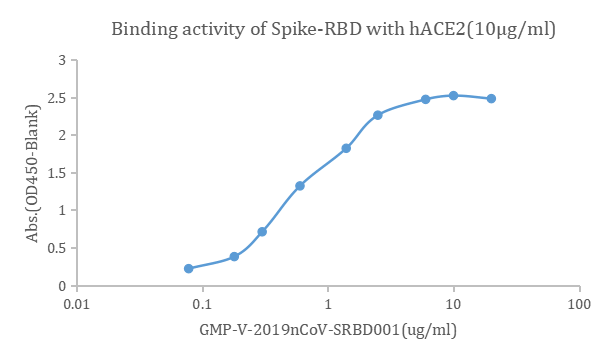
– Competition assay for neutralizing antibodies, peptides inhibitor and vaccines-immunized serums
Multi-VariantsTM SARS-CoV2 (wildtype, D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, and so on) Pseudotype virus-Based Neutralization Assay System for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies (Lentiviral pseudovirus)
Cat No. | Description | Order
|
|---|---|---|
Spike S943P mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
SARS-CoV-2 Pseudotyped virus packaging and production |
||
Spike D614G mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike V367F mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike G476S mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike V483A mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike H49Y mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike Q239K mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike A831V mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
||
Spike P1263L mutation SARS-CoV-2(2019nCoV) Pseudotyped virus |
GeneMedi SARS-CoV-2 Pseudovirus (PSV) Based Cell Entry
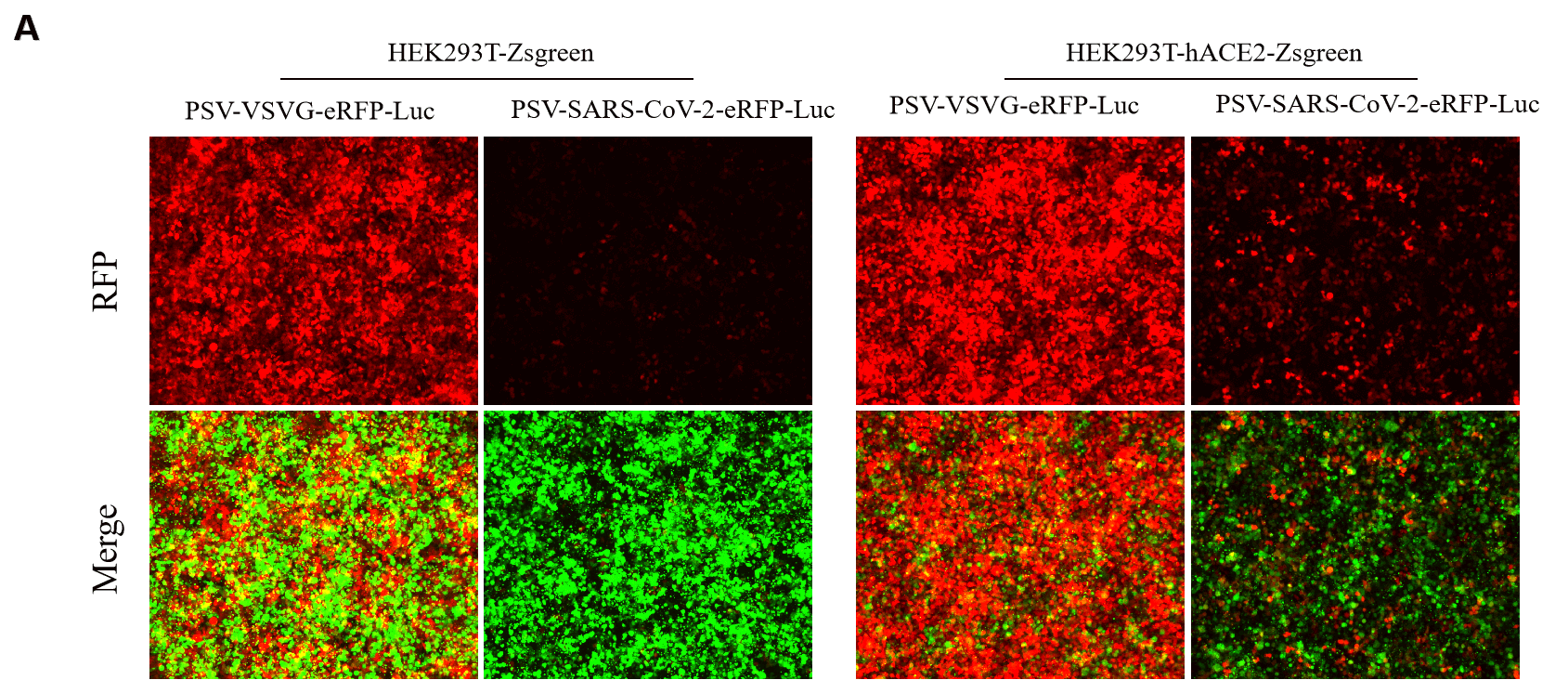
GeneMedi SARS-CoV-2 Pseudovirus (PSV) Based Cell Entry
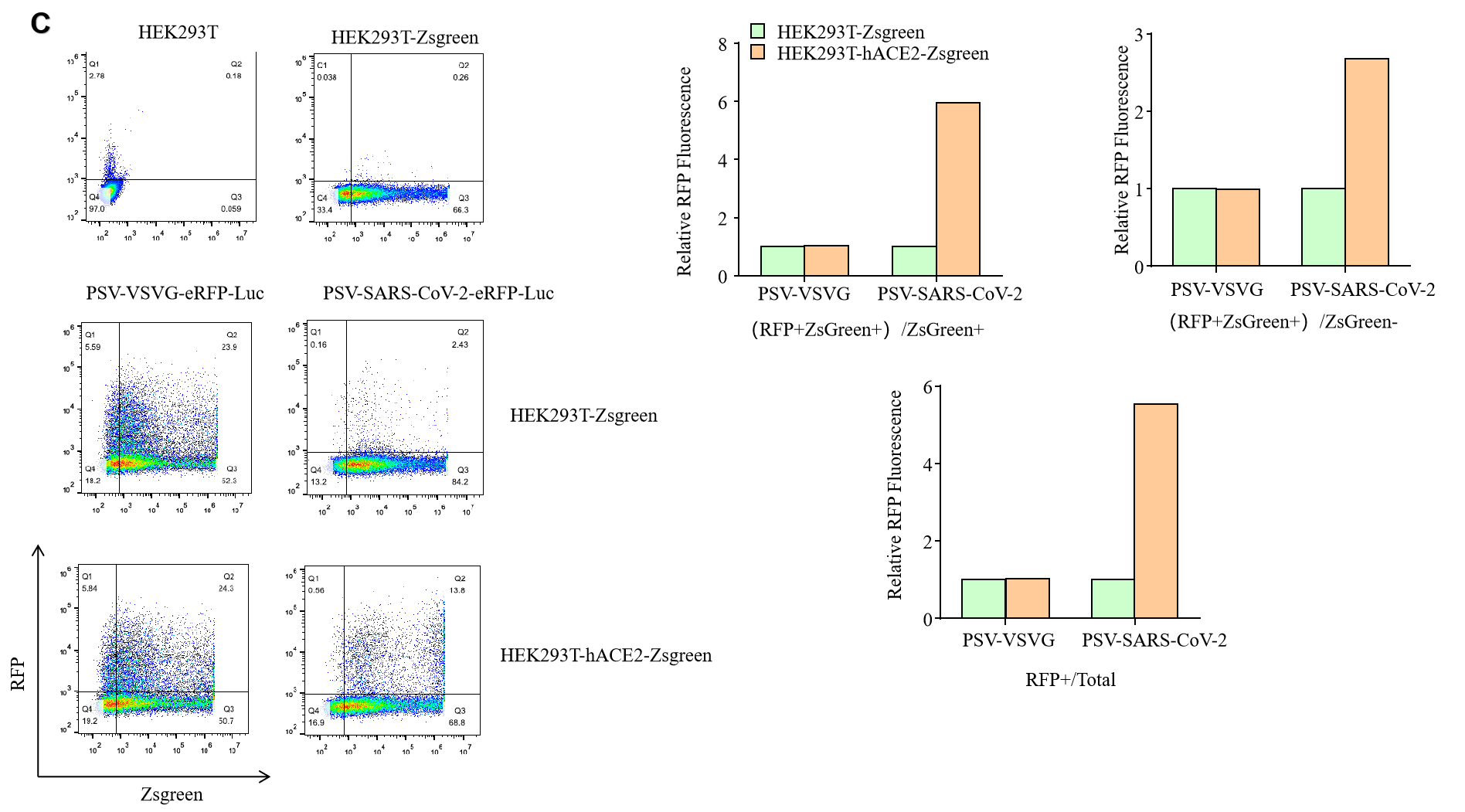
GeneMedi SARS-CoV-2 PSV-Luciferase (Cat.GM-2019nCoV-PSV01) is recombinant pseudotyped lentiviral particles containing SARS-CoV-2 spike protein to mimic SARS-CoV-2 (2019nCoV) cell infection.
GM-2019nCoV-PSV01 is a powerful tool for SARS-CoV-2 related vaccine efficacy evaluation, neutralizing antibodies, peptides blockers competitors neutralization assay, and tissue-specific infection determination.
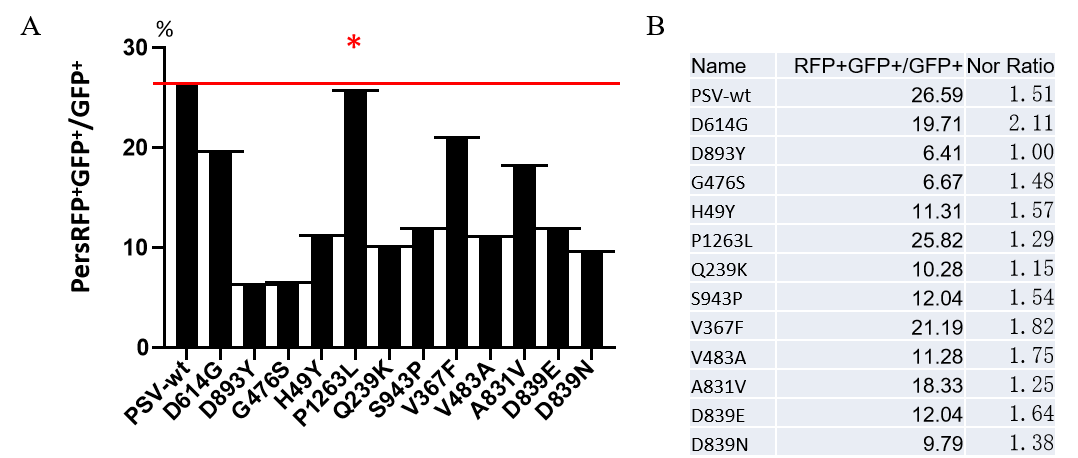 |
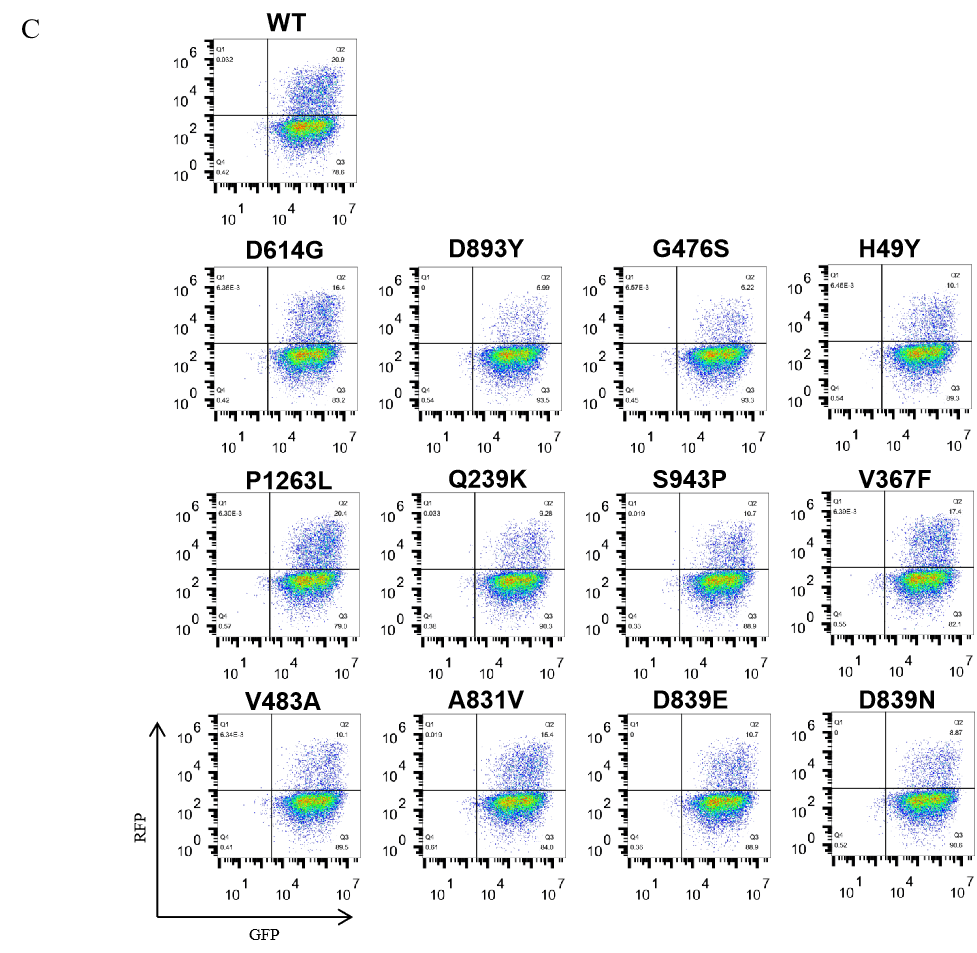
|
|
Figure. The Pseudovirus (PSV) Based Cell Entry assay was performed on 293T-hACE2 cells infected with GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants (D614G, S943P, V367F, G476S, V483A, H49Y, Q239K, A831V, P1263L, D839Y/N/E:D839Y, D839N, D839E) Pseudovirus (PSV) Infection rate was determined by RFP+GFP+/GFP+ with FACS validation. |
|
Codon-optimized mammalian expression vector for SARS-COV-2 (2019nCOV) spike wide type & mutant variants (D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, and so on)
Codon-optimized viral particle (Adenovirus, Lentivirus, AAV) for SARS-COV-2 (2019nCOV) spike wide type & mutant variants (D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, and so on)
Cat No. | Description | Vector | Reporter | Order
|
|---|---|---|---|---|
pGMLV-2019nCoV-spike(S1+S2,C-6His) |
Lentiviral vector |
Zsgreen |
||
pGMLV-2019nCoV-S1(C-6His) |
Lentiviral vector |
Zsgreen |
||
pGMLV-2019nCoV-Spike RBD(C-6His) |
Lentiviral vector |
Zsgreen |
||
pGMAAV-2019nCoV-Spike RBD(C-3FLAG) |
AAV vector |
Zsgreen |
||
pGMLV-2019nCoV-spike£¨S1+S2,C-6His£© |
Lentiviral vector |
Zsgreen |
||
Ad-2019nCoV-Spike(S1+S2,C-3FLAG) |
Pre-made adenovirus |
EGFP |
||
Ad-2019nCoV-Spike(S1 protein,C-3FLAG) |
Pre-made adenovirus |
EGFP |
||
Ad-2019nCoV-Spike(S protein RBD,C-3FLAG) |
Pre-made adenovirus |
EGFP |
||
Ad-2019nCoV-Spike(S1+S2,C-3FLAG) |
Pre-made adenovirus |
null |
||
Ad-2019nCoV-Spike D614G (S protein,S1+S2,D614G) |
Pre-made adenovirus |
null |
293T-ACE2 stable cell line & human ACE2 expression vectors: Effector cell of pseudotype virus-Based Neutralization Assay System for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies (Lentiviral pseudovirus)
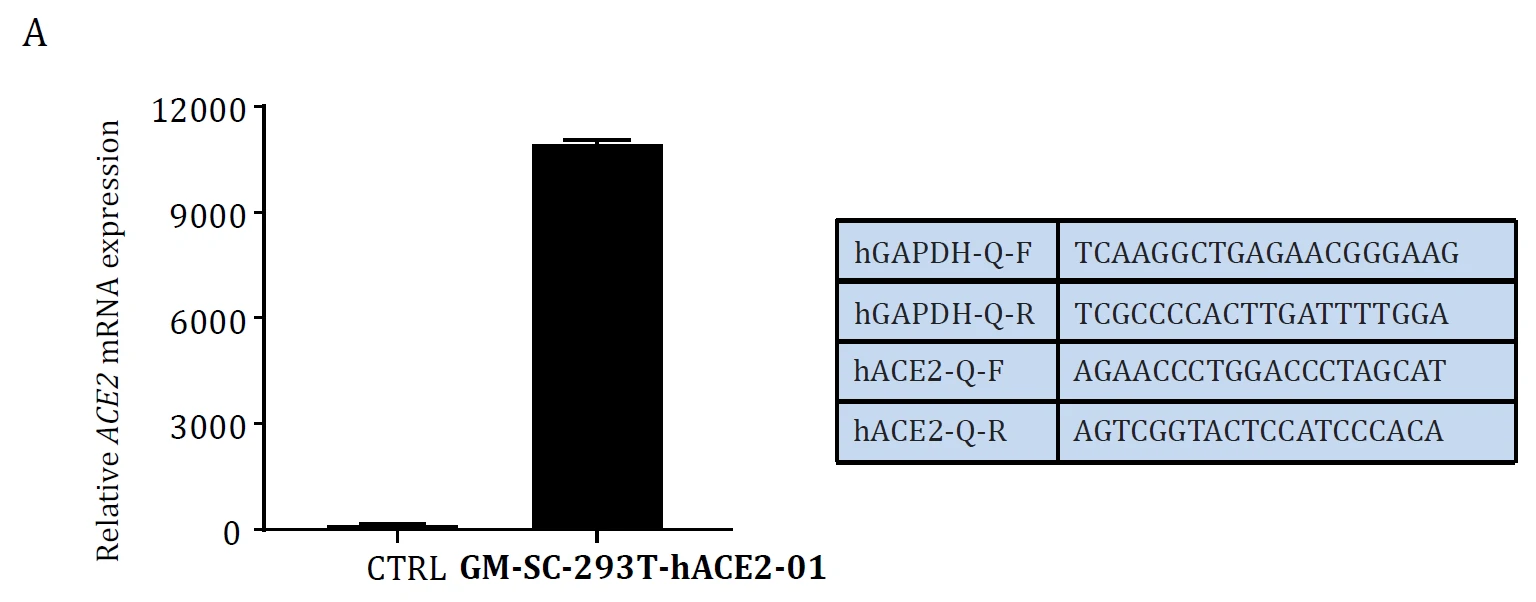 |
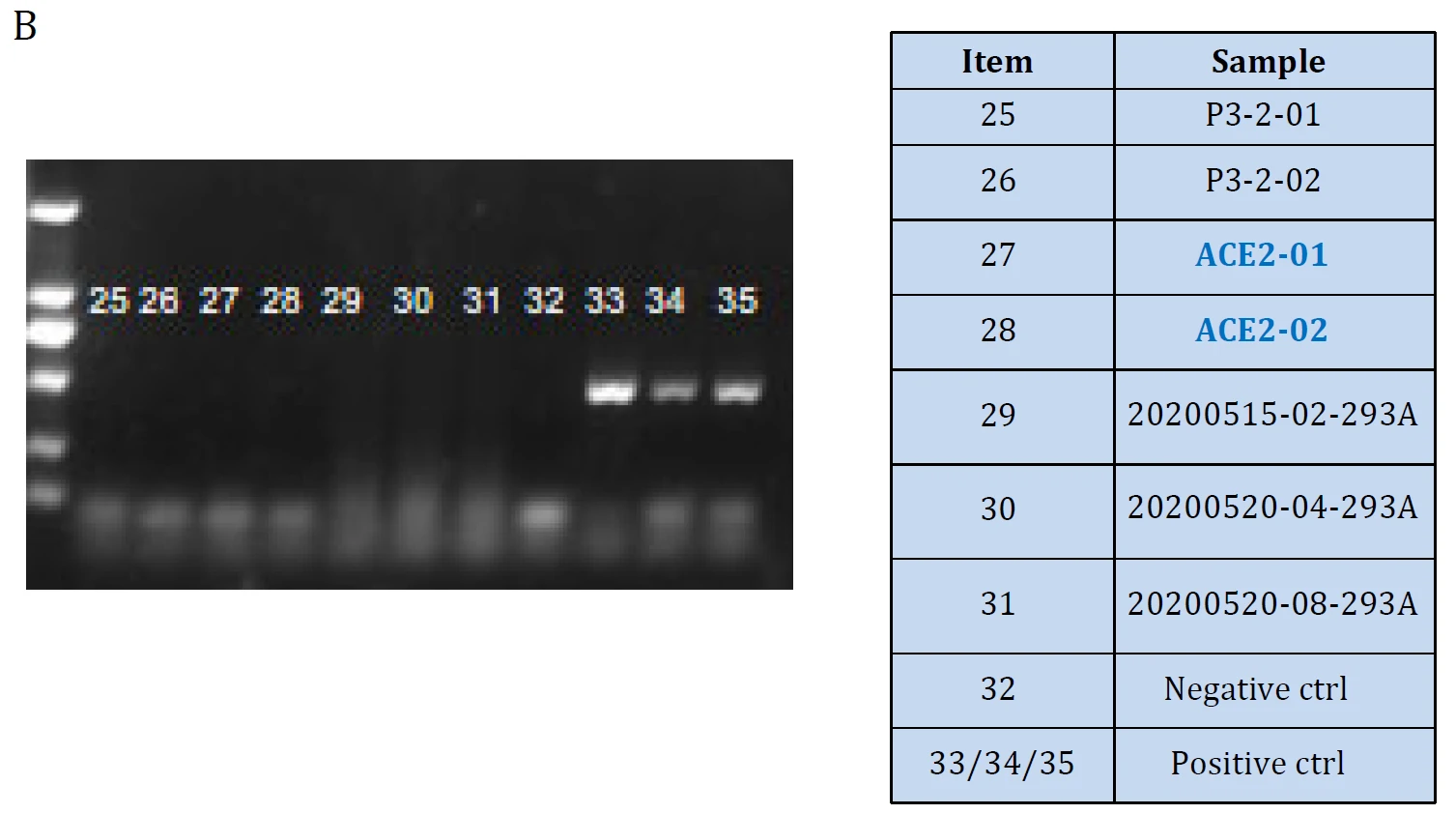
|
| Figure. ACE2 mRNA level Validation in hACE2 overexpression stable HEK293T cell lines: Cat. GM‐SC‐293T‐hACE2‐01 (A) and the cell lines were tested as Mycoplasma free (B). | |
| Cat No. | 2019 nCoV related Gene | Gene &Vector description of 2019 nCoV | Vector | Reporter | Tag | codon Optimized | Order |
| GMV-V-2019nCoV-041 | ACE2 | pGMLV-hACE2(C-3FLAG) | Lentiviral vector | Zsgreen | C-3FLAG | No |  |
| GMV-V-2019nCoV-042 | ACE2 | pAD-hACE2(C-3FLAG) | Pre-made Adenovirus | EGFP | C-3FLAG | No |  |
| GMV-V-2019nCoV-043 | ACE2 | pAD-mACE2(C-3FLAG) | Pre-made Adenovirus | EGFP | C-3FLAG | No |  |
| GMV-V-2019nCoV-044 | ACE2 | pGMLV-mACE2(C-3FLAG) | Lentiviral vector | Zsgreen | C-3FLAG | No |  |
Validated SARS-CoV-2 neutralizing antibodies-benchmark COVID-19 neutralizing antibodies.
| Cat No. | Antigen Name of 2019-nCoV(SARS-CoV-2) | Source (Expression Host) | Isotypes | Bioactivity validation | Order |
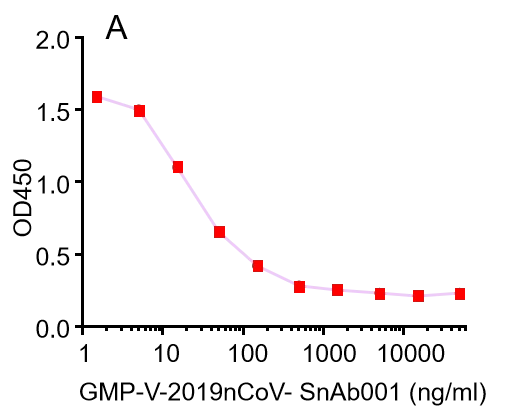
| GMP-V-2019nCoV-SnAb001 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1) | Mammalian (human cell) | human IgG1 | Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity. COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |  |
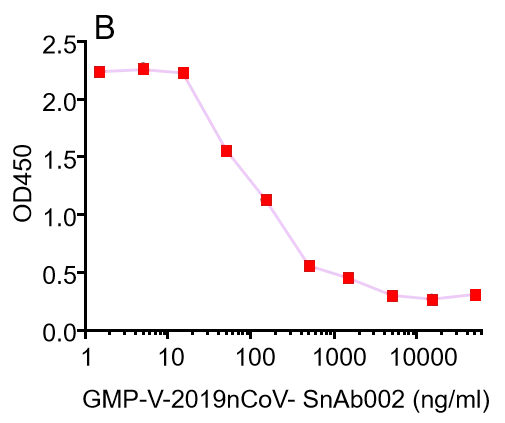
| GMP-V-2019nCoV-SnAb002 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM) | Mammalian (human cell) | human IgM | Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity. COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |  |
| GMP-V-2019nCoV-SnAb003 | Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA) | Mammalian (human cell) | human IgA | Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity. COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |  |
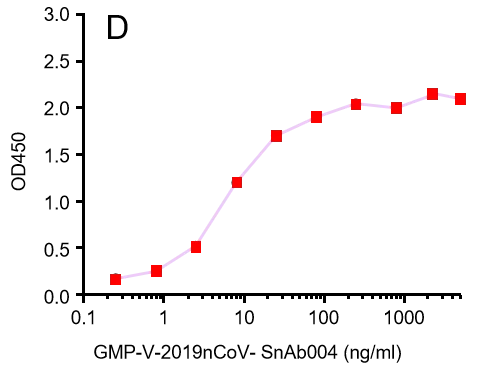
| GMP-V-2019nCoV-SnAb004 | Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1) | Mammalian (human cell) | mouse IgG1 | Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity. COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |  |
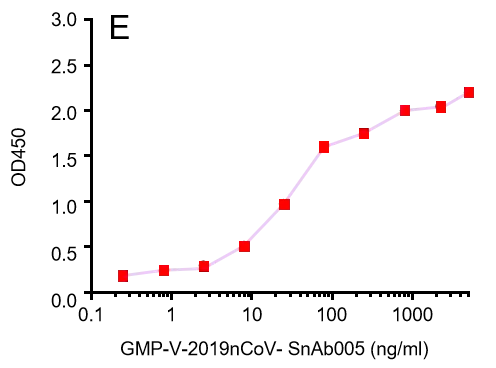
| GMP-V-2019nCoV-SnAb005 | Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1) | Mammalian (human cell) | Cynomolgus (Non human primate, NHP) IgG1 | Validated in COVID-19 Spike protein and Spike-RBD protein binding affnity. COVID-19 related neutralizing potency is validated by 1.2019nCoV pseudotyped virus based neutralization assay in 293T-ACE2 effector cell. 2. competitively blocking the binding of ACE-2 receptor with SARS-CoV-2 Spike protein. |  |
 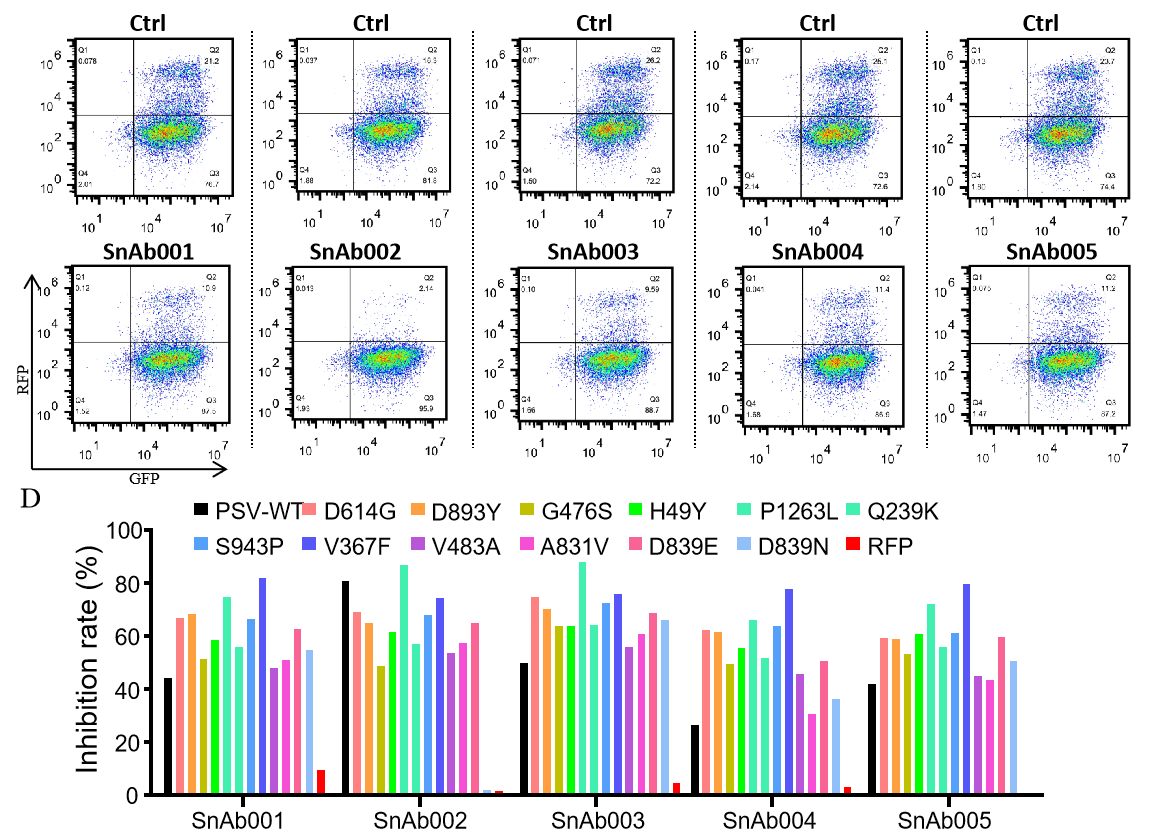
|
| Figure. The Pseudovirus (PSV) Based Neutralizing Assay was performed on 293T-hACE2 cells infected with GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants (D614G, S943P, V367F, G476S, V483A, H49Y, Q239K, A831V, P1263L, D839Y/N/E:D839Y, D839N, D839E) Pseudovirus (PSV) under treatment of GeneMedi’s anti-2019-nCoV Spike Neutralizing antibodies (Nabs) . Inhibition rate was determined by comparing the relative RFP+GFP+/GFP+ rate. |
GeneMedi’s anti-2019-nCoV Spike Neutralizing antibodies (Nabs) and Spike RBD protein binding validation
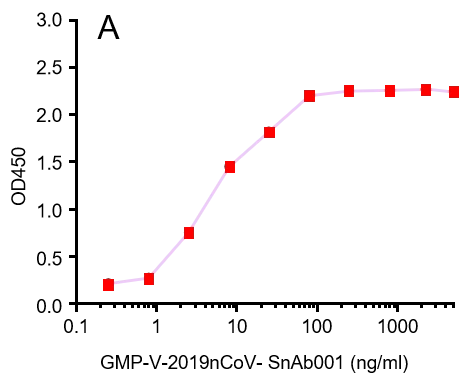
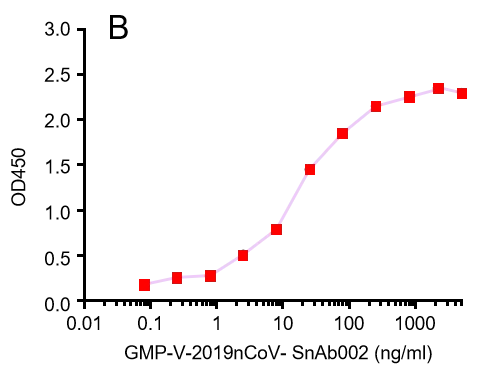
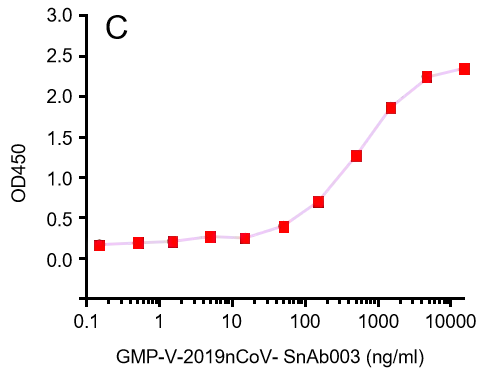
A.
GMP-V-2019nCoV-SnAb001:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1)
B.
GMP-V-2019nCoV-SnAb002:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM)
C.
GMP-V-2019nCoV-SnAb003:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA)
D.
GMP-V-2019nCoV-SnAb004:Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1)
E.
GMP-V-2019nCoV-SnAb005:Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1)
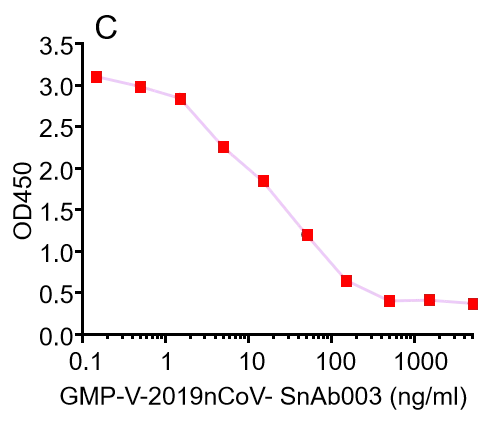
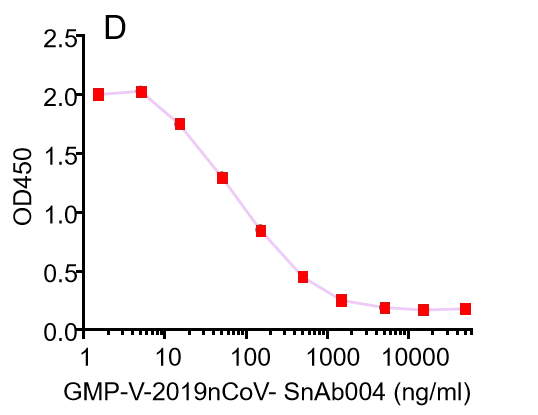
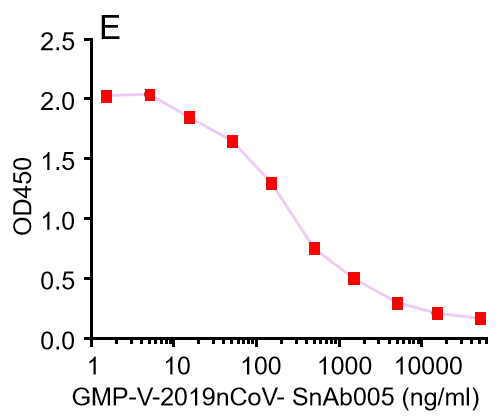
A.
GMP-V-2019nCoV-SnAb001:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgG1)
B.
GMP-V-2019nCoV-SnAb002:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgM)
C.
GMP-V-2019nCoV-SnAb003:Anti-2019-nCoV Spike (Spike RBD domain) human monoclonal neutralizing antibody (IgA)
D.
GMP-V-2019nCoV-SnAb004:Anti-2019-nCoV Spike (Spike RBD domain) mouse monoclonal neutralizing antibody (IgG1)
E.
GMP-V-2019nCoV-SnAb005:Anti-2019-nCoV Spike (Spike RBD domain) Cynomolgus monoclonal neutralizing antibody (IgG1)
GeneMedi’s efficacy evaluation solutions will be useful for your anti-COVID-19 candidates funtional assay and evaluation,which is including:
1) Types of Vaccines (by testing immunized serum from human, mouse, NHP etc.)
2) Neutralizing antibodies
3) Peptides blockers (peptide inhibitors)
4) Compounds targeting Spike induced cell-fusion.
INFORMATION COLLECTION
Landscape of global COVID-19 vaccine candidates development
COVID-19 Guidance and information collection on the new mutant variants of the SARS-CoV-2 (2019nCoV) virus
– New SARS-COV-2 variant: information and risk assessment
– Central Alerting System (CAS) alert
– New variant clustering in households analysis (ONS)
– SARS-CoV-2 lateral flow antigen tests: evaluation of VUI-202012/01
– WTO: Statement of the WHO Working Group on COVID-19 Animal Models (WHO-COM) about the UK and South African SARS-CoV-2 new variants.
– EMA guidance for COVID-19 vaccine
– Investigation of novel SARS-CoV-2 variant: Variant of Concern 202012/01
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 6
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 5
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 4
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 3
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 2
• Investigation of novel SARS-CoV-2 variant: 202012/01. Technical briefing 1
New variant of SARS-COV-2 (2019nCOV) B.1.1.7 lineage spreaded in UK
The world is in midst of the COVID-19 pandemic. Recently a novel SARS-COV-2 (2019nCOV) lineage, the B.1.1.7 lineage, with serials of site mutation, shows stronger infection ability in the UK. The SARS-COV-2 B.1.1.7 lineage carries a larger than a usual number of coronavirus genetic changes.
Extended Reading: Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations
Relative products collection:
GeneMedi products for New variant of SARS-COV-2 (2019nCOV) UK B.1.1.7 lineage
New variant of SARS-COV-2 (2019nCOV) 501Y.V2 lineage(B.1.351) spreaded in South African SARS-CoV-2
The South African variant is characterized by eight lineage-defining mutations in the spike protein including three key residues in the receptor binding domain (K417N, E484K and N501Y) and is referred to as lineage 501Y.V2.
Extended Reading: Alert Notification: New SARS-CoV-2 variant with multiple spike protein mutations
Relative products collection:
GeneMedi products for New variant of SARS-COV-2 (2019nCOV) South Africa 501Y.V2 lineage(B.1.351)
COVID-19 News and announcements collection on the new mutant variants of the SARS-CoV-2 (2019nCoV) virus
– Rapid evaluation confirms lateral flow devices effective in detecting new COVID-19 variant
– Confirmed cases of COVID-19 variant from South Africa identified in UK
– Statement from Chief Medical Officer, Professor Chris Whitty, about new strain of COVID-19
– NERVTAG statements on COVID-19 (SARS-CoV-2)
• NERVTAG/SPI-M Extraordinary meetingon SARS-CoV-2 variant of concern 202012/01 (variant B.1.1.7)
• NERVTAG meetingon SARS-CoV-2 variant under investigation VUI-202012/01
• CPR as an AGP – Evidence review and NERVTAG consensus
About COVID-19 Pandemic and SARS-CoV-2 Vaccine
Coronavirus Disease 2019 (COVID-19) is a novel viral pneumonia caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). First discovered in Wuhan, a city in Hubei province of China, COVID-19 has already broken out throughout the world and posed a great threat to the public health, especially in Europe and North America now. Additionally, person-to-person transmission of COVID-19 disease is reported to be extremely rapid [158-160]. To date, more than one million cases were infected with COVID-19 and over 55,000 deaths occurred. Therefore, it is really urgent and noteworthy to develop the vaccines specific to COVID-19/SARS-CoV-2.
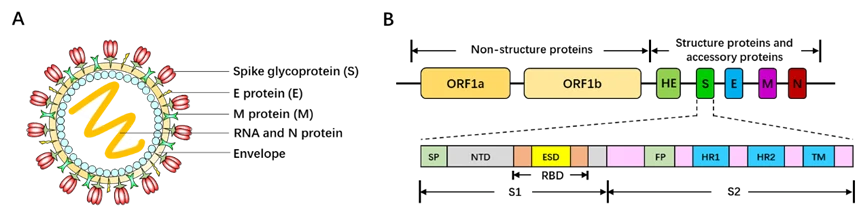
Belonging to the Betacoronavirus genus family, SARS-CoV-2 is 60~200nm in diameter and encapsidates a large positive-sense, single-stranded RNA virus (26-32kb) with many spikes on the virus capsid (Fig. 17A). The RNA genome of SARS-CoV-2 encodes several accessory proteins and structural proteins, such as nucleocapsid (N) protein, envelope (E) protein, membrane (M) protein, and spike (S) protein (Fig. 10B). Although the detailed mechanism of SARS-CoV-2 infection has not been clearly illuminated, several studies demonstrated that SARS-CoV-2 enters human cells via utilizing spike (S) protein to bind to the angiotensin converting enzyme (ACE2) on the surface of target cell [161, 162].
Since the genome sequences of SARS-CoV were discovered and reported (https://www.gisaid.org/CoV2020/), a large number of pharmaceutical enterprises and research organizations are sparing all efforts to the vaccine development. Different companies utilize different targets and antigen epitopes. Some of the advances are listed in the following Table 10 (from WHO), and most of them focus on viral vector-based vaccines (replicating or non-replicating viral vector-based vaccines), recombinant protein (Spike), and nucleic acid-based vaccines. To date, two COVID-19 vaccines have entered Phase I clinical testing to assess the safety and potency of vaccines. One is mRNA-1273, was developed by Moderna Therapeutics, encoding a prefusion-stabilized form of Spike (S) protein [163] (https://www.nature.com/articles/d41587-020-00005-z). Another vaccine is recombinant protein of SARS-CoV-2 antigen, developed by Chinese Academy of Military Sciences, Institute of Military Medicine. It was predicted that these vaccines can be applied in clinics in a large scale as early as 2021 if they can successfully pass the clinical testing. Although there is a long way for theses vaccines to be applied for prevention and therapy of COVID-19, they indeed bring great hope and light to people all over the world.
References
1. 2 Korber, B. et al. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv Preprint., doi:10.1101/2020.04.29.069054 (2020).
2. Investigation of novel SARS-COV-2 variant. Public health England.
3. European Centre for Disease Prevention and Control. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom – 20 December 2020.ECDC: Stockholm; 2020.
4. Li, G. & De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews Drug Discovery, doi:10.1038/d41573-020-00016-0 (2020).
5. Haque, A. & Pant, A. B. Efforts at COVID-19 Vaccine Development: Challenges and Successes. Vaccines 8, doi:10.3390/vaccines8040739 (2020).
6. Dong, Y. et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduction and Targeted Therapy 5, doi:10.1038/s41392-020-00352-y (2020).
7. Andrew Rambaut1, N. L., Oliver Pybus, Wendy Barclay, Jeff Barrett5, Alesandro Carabelli6, Tom Connor, Tom Peacock, David L Robertson8, Erik Volz,. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/ (2020).
8. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. European Centre for Disease Prevention and Control: Publications & data.
9. Zhang, L. et al. SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome. bioRxiv Preprint. , doi:10.1101/2020.12.12.422516 (2020).