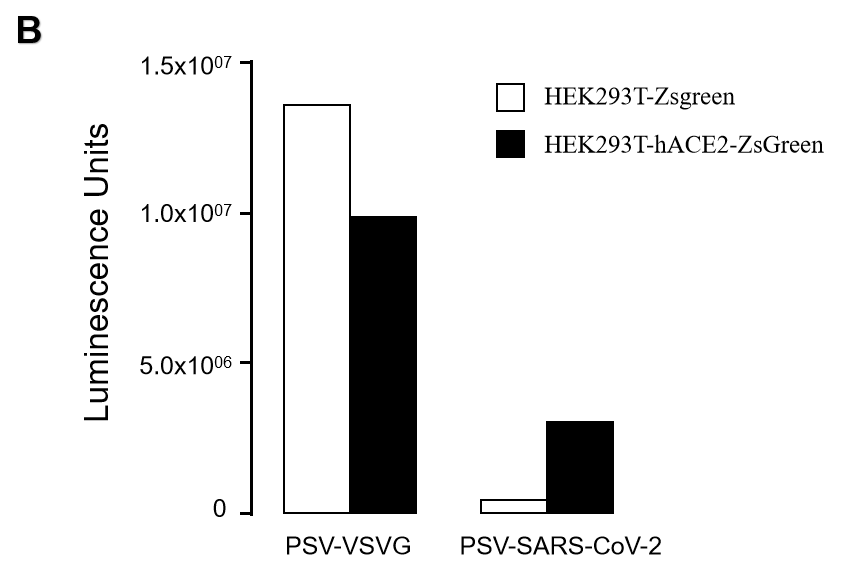SARS-CoV2 (wildtype 2019nCoV, D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2?lineage etc.) Coronavirus Pseudotype virus-Based Neutralization Assay System for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies (Lentiviral pseudovirus)
Diagnostic antibodies and antigens for Companion Animal disease testing
● Rabbit
Diagnostic antibodies and antigens for Swine disease testing
Diagnostic antibodies and antigens for Avian disease testing
Diagnostic antibodies and antigens for Multiple animal disease testing
Diagnostic antibodies and antigens for Ruminant disease testing
● Deer
Diagnostic antibodies and antigens for infectious and non-infectious Equine/Horse disease testing
SOCAIL MEDIA

Content index
COVID-19, Why GeneMedi's Multi-VariantsTM SARS-CoV-2 Pseudovirus Based Neutralization Assay system?
The outbreak of COVID-19, caused by SARS-CoV-2 (2019-nCoV), has been a global pandemic and cause millions of people dead. The coronavirus is kept accumulated mutating. The most important mutation occurs in SARS-CoV-2 (2019nCoV) Spike protein (SARS-CoV-2 S protein). The SARS-CoV-2 (2019nCoV) Spike mediates binding and entry into host cells and is a major target of neutralizing antibodies1.
The United Kingdom (UK) has faced a rapid increase in COVID-19 cases caused by a novel SARS-COV-2 (2019nCOV) lineage, the B.1.1.7 lineage, which carries a larger than a usual number of coronavirus genetic changes2-3, particularly in the SARS-CoV-2 spike protein. The South African variant is characterized by eight lineage-defining mutations in the spike protein including three key residues in the receptor-binding domain (K417N, E484K, and N501Y) and is referred to as lineage 501Y.V2.
Due to its high pathogenicity and infectivity1, live SARS-CoV-2 should be handled under biosafety level 3 (BSL-3) conditions. GeneMedi has developed wild-type and mutant variants of the SARS-CoV-2 pseudovirus assay system, from which the SARS-CoV-2 pseudotyped virus can be handled in biosafety level 2 (BSL-2)4. GeneMedi’s Multi-VariantsTM SARS-CoV2 (wildtype 2019nCoV, D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, etc.) pseudotype virus-based neutralization assay system is a powerful tool of SARS-CoV-2 for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies (Lentiviral pseudovirus), which is kept updating by tracking the newest variants.
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) UK B.1.1.7 lineage and South Africa 501Y.V2 lineage, and Brazilian P.1 lineage(B.1.1.28.1)
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) UK B.1.1.7 lineage
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) B.1.1.7 lineage, which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Other products of SARS-COV-2 (2019nCOV) B.1.1.7 lineage:
1) SARS-COV-2 (2019nCOV) B.1.1.7 lineage of Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
2) GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) B.1.1.7 lineage
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) B.1.1.7 lineage.
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) 501Y.V2 lineage
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) S501Y.V2 lineage, which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Other products of SARS-COV-2 (2019nCOV) South Africa 501Y.V2 lineage, B.1.351 :
1) SARS-COV-2 (2019nCOV) – South African variant 501Y.V2 lineage, B.1.35 of Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
2) GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) S501Y.V2 lineage
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) S501Y.V2 lineage.
GeneMedi pseudotyped virus (pseudovirus) of SARS-COV-2 (2019nCOV) Brazilian variants: P.1 lineage(B.1.1.28.1)
Taking responsibility to help accelerate the COVID-19 vaccine and therapeutic antibody discovery and development, GeneMedi had developed the pseudotype virus (pseudovirus) of SARS-COV-2 (2019nCOV) P.1 lineage(B.1.1.28.1, Brazilian variant), which will meet the evaluation of the efficacy of COVID19 vaccines and therapeutic antibodies.
Other products of SARS-COV-2 (2019nCOV) Brazilian variants: P.1 lineage(B.1.1.28.1) :
1) SARS-COV-2 (2019nCOV)-Brazilian variants: P.1 lineage(B.1.1.28.1) of Spike protein & ACE2 competition binding assay for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies
2) GeneMedi codon-optimized spike mammalian expression vector for SARS-COV-2 (2019nCOV) Brazilian variants: P.1 lineage(B.1.1.28.1)
GeneMedi designed a mammalian expression codon-optimized spike mutation/deletion variant vector for COVID-19 SARS-COV-2 (2019nCOV) P.1 lineage(B.1.1.28.1, Brazilian variant)
GeneMedi Multi-VariantsTM SARS-CoV-2(2019nCoV) Pseudovirus Based Neutralization Assay system5
The Multi-VariantsTM SARS-CoV2 (2019nCoV) coronavirus Pseudotype virus-Based Neutralization Assay System a powerful tool designed for efficacy evaluation of COVID-19 vaccines and therapeutic antibodies.
The system is including:
1. Multi-VariantsTM SARS-CoV2 (wildtype, D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, etc.) Pseudotype virus (Lentiviral pseudovirus)
2. 293T-ACE2 stable cell line & human ACE2 expression vectors: Effector cell of pseudotype virus-Based Neutralization Assay System.
3. Validated SARS-CoV-2 neutralizing antibodies-benchmark COVID-19 neutralizing antibodies.
4. Multi-VariantsTM Codon-optimized adenoviral vector (adenovirus particle) for SARS-COV-2 (2019nCOV) spike wide type & mutant variants (D614G, N501Y, E484K, B.1.1.7 lineage, 501Y.V2 lineage, etc.)
SARS-CoV2 Pseudotype Virus (Lentiviral pseudovirus particle)
GeneMedi’s Multi-VariantsTM SARS-CoV-2 Pseudovirus particle with luciferase/RFP double reporters (Cat.GM-2019nCoV-PSV) is recombinant pseudotyped lentiviral particles combining chimeric SARS-CoV-2 spike protein envelop to mimic SARS-CoV-2 (2019nCoV) cell infection and cell entry5.
The SARS-CoV-2 pseudovirus particles encode firefly luciferase and RFP in their lentiviral vector genome. The firefly luciferase and RFP gene will be strongly expressed after the SARS-CoV-2 pseudovirus entry into ACE2-expressing cells. Actually, 293T-hACE2(human ACE2 overexpression stable HEK293T cell lines) is normally used as an effector cell.
GM-2019nCoV-PSV is a powerful tool for SARS-CoV-2 related vaccine candidates’ efficacy evaluation, neutralizing antibodies, peptides blockers competitors neutralization assay, and tissue-specific infection determination.
293T-ACE2 stable cell line & human ACE2 expression vectors: Effector cell of pseudotype virus-Based Neutralization Assay System
 |

|
|
Figure. ACE2 mRNA level Validation in hACE2 overexpression stable HEK293T cell lines: Cat. GM‐SC‐293T‐hACE2‐01 (A) and the cell lines were tested as Mycoplasma free (B). |
|
Validation Data Post: Multi-VariantsTM SARS-CoV-2(2019nCoV) Pseudotyped Virus Based Neutralization Assay5
In Multi-VariantsTM Pseudovirus Based Neutralization Assay (PBNA), the inhibition of viral entry into cells by neutralizing antibodies is correlated to the decreased levels of firefly luciferase signals in the ACE2 expression cells (hACE2-HEK293T).
GeneMedi’s Multi-VariantsTM Pseudovirus Based Neutralization Assay (PBNA) is a conventional assay method that is suitable for High-Throughout Screening (HTS) without live virus engaged.
GeneMedi’s Multi-VariantsTM Pseudotype virus-Based Neutralization Assay System is a powerful tool for efficacy evaluation assay of your anti-COVID-19 candidate, which is including:
1) Types of Vaccines (by testing immunized serum from human, mouse, NHP, etc.)
2) Neutralizing antibodies
3) Peptides blockers (peptide inhibitors)
4) Compounds targeting Spike-induced cell-fusion.
GeneMedi SARS-CoV-2 Pseudovirus (PSV) Based Cell Entry
GeneMedi SARS-CoV-2 Pseudovirus (PSV) Based Cell Entry
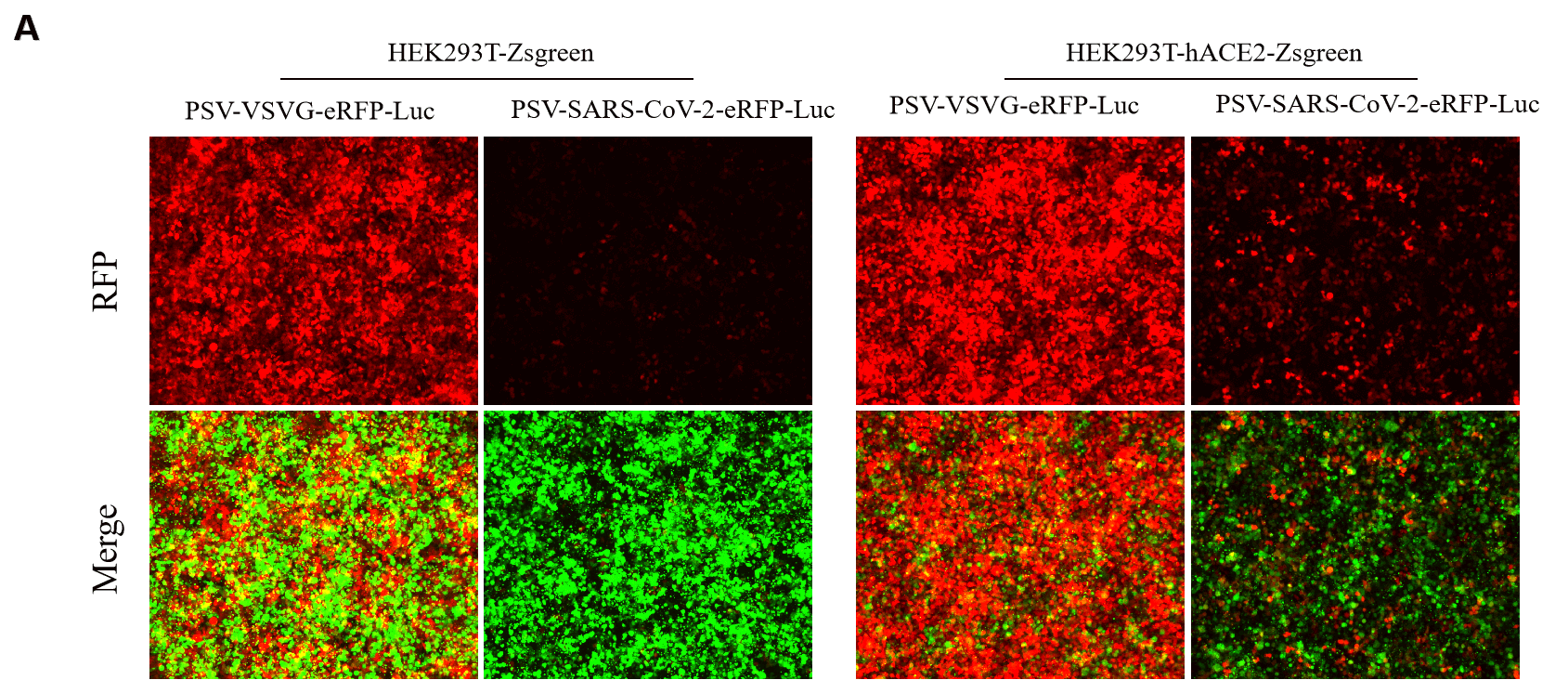
GeneMedi SARS-CoV-2 PSV-Luciferase (Cat.GM-2019nCoV-PSV01) is recombinant pseudotyped lentiviral particles containing SARS-CoV-2 spike protein to mimic SARS-CoV-2 (2019nCoV) cell infection.
GM-2019nCoV-PSV01 is a powerful tool for SARS-CoV-2 related vaccine efficacy evaluation, neutralizing antibodies, peptides blockers competitors neutralization assay, and tissue-specific infection determination.
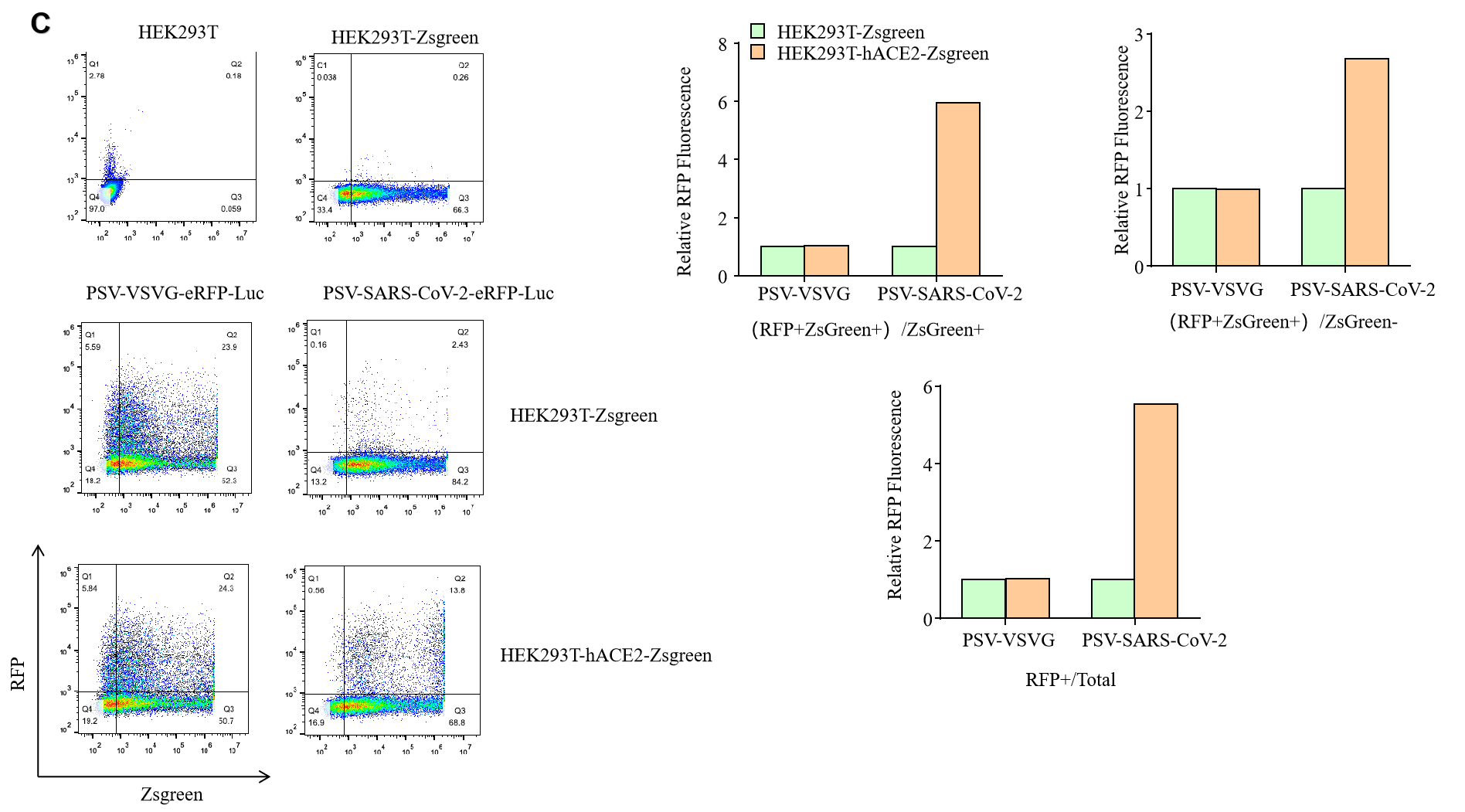
 |
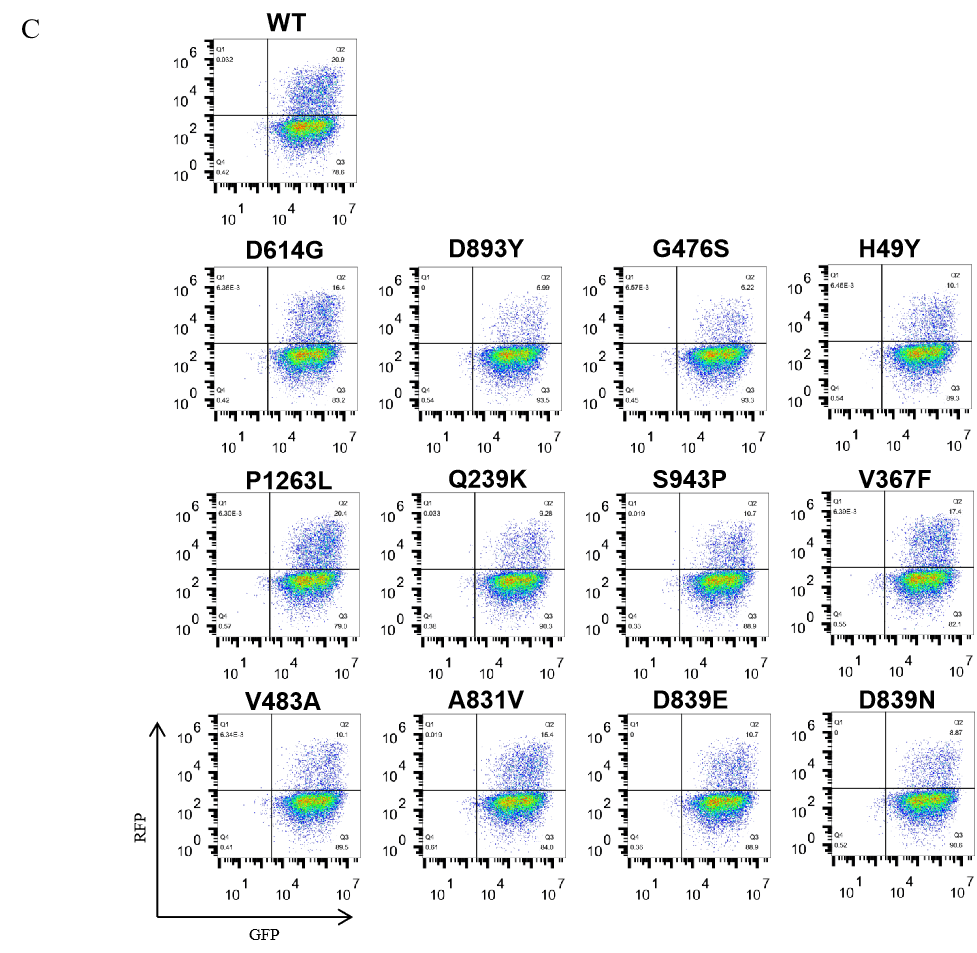
|
|
Figure. The Pseudovirus (PSV) Based Cell Entry assay was performed on 293T-hACE2 cells infected with GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants (D614G, S943P, V367F, G476S, V483A, H49Y, Q239K, A831V, P1263L, D839Y/N/E:D839Y, D839N, D839E) Pseudovirus (PSV) Infection rate was determined by RFP+GFP+/GFP+ with FACS validation. |
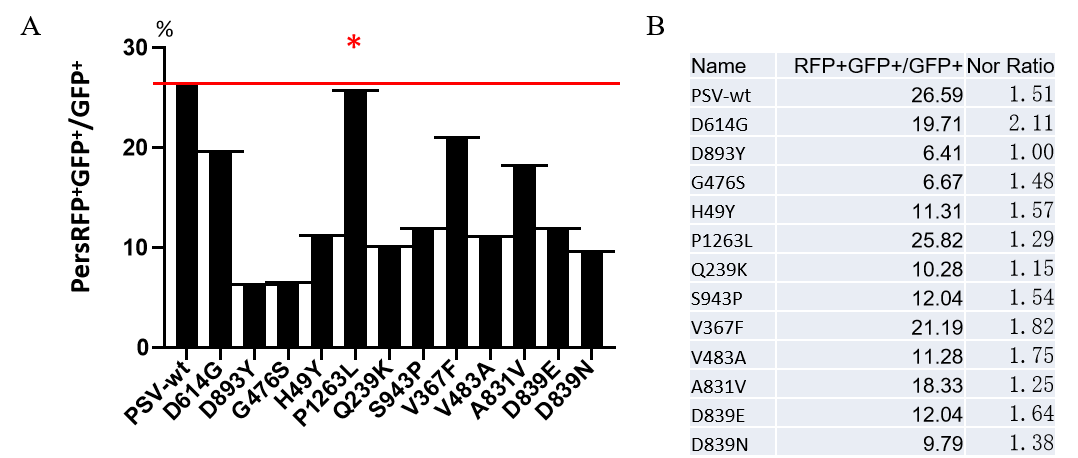
|
 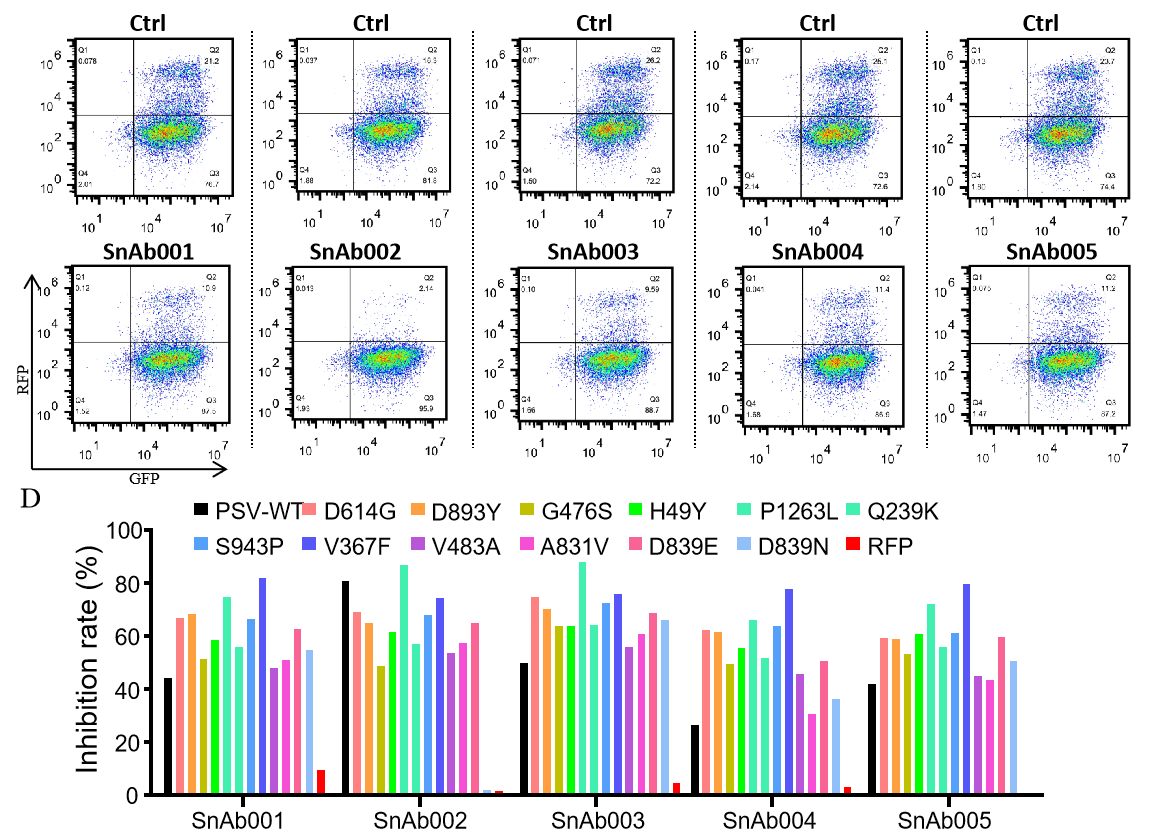
|
|
Figure. The Pseudovirus (PSV) Based Neutralizing Assay was performed on 293T-hACE2 cells infected with GeneMedi-SARS-CoV-2 WT and Spike Mutation Variants (D614G, S943P, V367F, G476S, V483A, H49Y, Q239K, A831V, P1263L, D839Y/N/E:D839Y, D839N, D839E) Pseudovirus (PSV) under treatment of GeneMedi’s anti-2019-nCoV Spike Neutralizing antibodies (Nabs) . Inhibition rate was determined by comparing the relative RFP+GFP+/GFP+ rate. |
References
2. Andrew Rambaut1, N. L., Oliver Pybus, Wendy Barclay, Jeff Barrett5, Alesandro Carabelli6, Tom Connor, Tom Peacock, David L Robertson8, Erik Volz, . Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/ (2020).
3. Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. European Centre for Disease Prevention and Control: Publications & data.
4. Jean K. Millet1, Tiffany Tang3, Lakshmi Nathan3, Javier A. Jaimes4, Hung-Lun Hsu3,5, & Susan Daniel3, G. R. W. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J Vis Exp, doi:10.3791/59010 (2019).
5. Nie, J. et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9, 680-686, doi:10.1080/22221751.2020.1743767 (2020).